Recently, a research team from Huazhong Agricultural University (HZAU) online published a review entitled Pasteurella multocida: Genotypes and Genomics in Microbiology and Molecular Biology Reviews, a journal on microbiology. Based on a large number of experimental data and analysis, this review systematically discusses their progress on P. multocida genome.

P. multocida which displays host specificity is a highly versatile pathogen capable of causing infections in humans and nonhuman primates. For a long time, due to the lack of fast and accurate bacterial typing technology and genetic operation system, the understanding of the pathogenic serotype and genotype of P.multocida and the molecular mechanism of host specificity is still unclear, and the genetic basis for determining P.multocida phylogenetics also needs to be further elucidated.
In this review, The team defined the dominant genotypes and the molecular biological characteristics of P. multocida from different host by applying genotyping into P. multocida and clarified the molecular basis for determining host preference of P. multocida. They proposed lipopolysaccharide (LPS) genotype and Multiple sequence locus genotype is the main determinant of the occurrence of P. multocida system. Professor Wu Bin from HZAU and Professor Brenda A. Wilson from University of Illinois at Urbana-Champaign(UIUC) are the corresponding authors of the review. Postdoc fellow Peng Zhong is the first author. Academician Chen Huanchun and other professors offered their suggestions for revision.
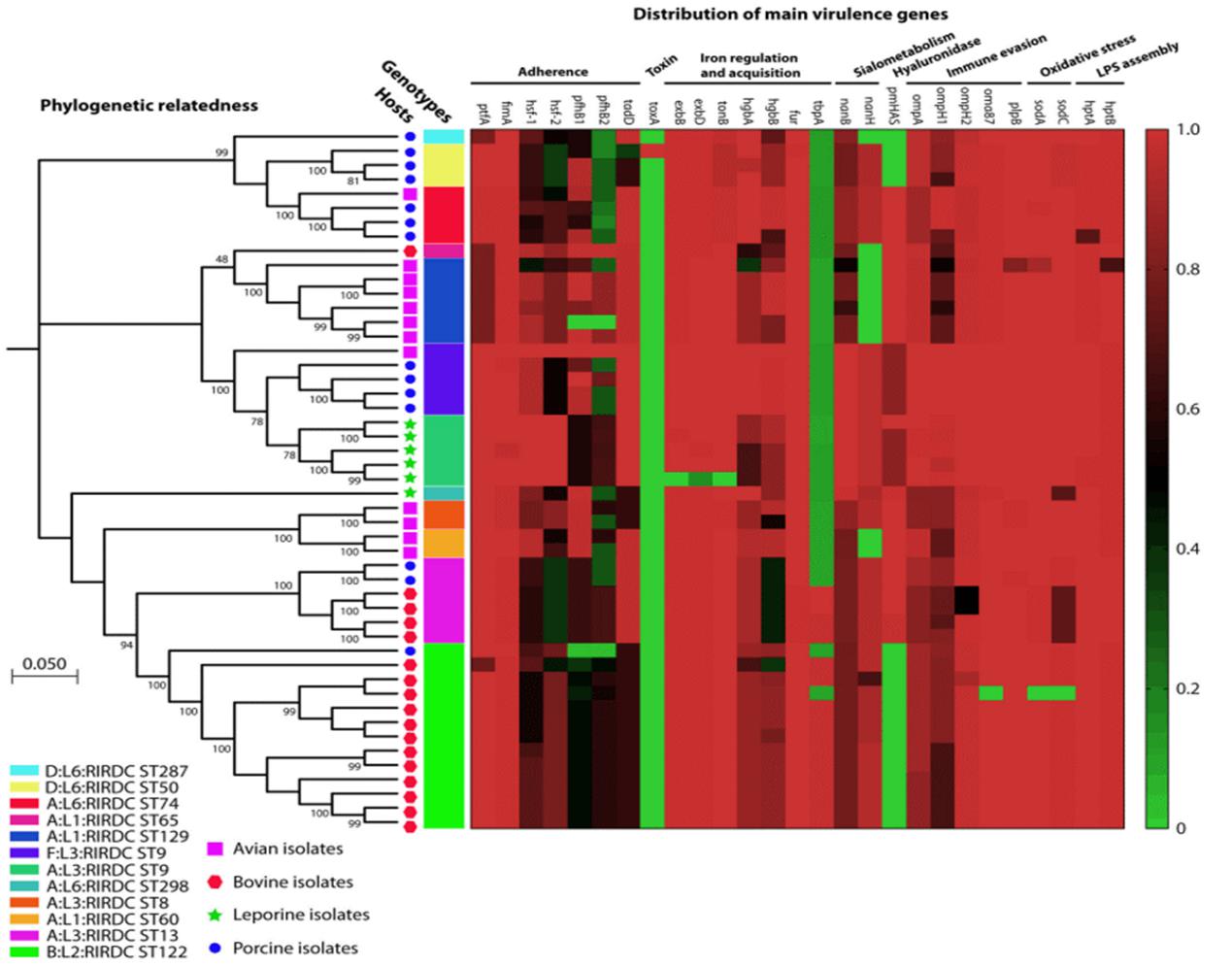
Distribution of the Main Virulence Genes Found among P. multocida Isolates of Different Genotypes and/or from Different Host Species
In recent years, the team has conducted research on pathogenesis, epidemiology and genomics of P. multocida. Their findings have been published in Front Microbiol, Arch Microbiol, Arch Microbiol, Vet Microbiol and Gene.
Source: http://news.hzau.edu.cn/2019/0905/54843.shtml
Translated by: Hou Yuanyuan
Supervised by: Guo Haiyan
